Our Research
Aim

The aim of the research in our lab is to understand the conformational switches of proteins with particular emhpasis on amyloid disease, protein-protein interactions and enzymes. We use a variety of techniques to solve the structures and dynamics of proteins under action and to study the conformational transition of amyloidogenic proteins including as a major experimental tool Nuclear Magnetic Resonance Spectroscopy (NMR).
Amyloids in Health and Disease
Alzheimer and prion diseases belong to amyloid diseases for which a normal host polypeptide accumulates in an abnormal beta-sheet -rich form in fibrillar aggregates. In prion diseases, the mostly alpha-helical host prion protein undergoes a conformational switch to a structure with enhanced beta-sheet content. It is believed that this converted abnormal form of the prion protein is the infectious agent of bovine spongiform encephalopathy (BSE), scrapie in sheep and Creutzfeldt-Jakob disease (CJD) in humans. In Alzheimer's disease, a peptide with mostly 'random-coil' like structure is converted to a beta-sheet-rich polypeptide clustered in fibrils. These fibrils are toxic to cells. Furthermore, the phenotypes of the well characterized yeast prions [PSI] and [URE3] and the prion phenotype [HET-s] in Podospora anserina are associated with amyloid formation. Since in all mentioned systems the polypeptides involved undergo a structural transition, detailed knowledge of the structures and dynamics of the normal and abnormal conformers and of the conformational transitions themselves are important aims of our work.
Nuclear Magnetic Resonance
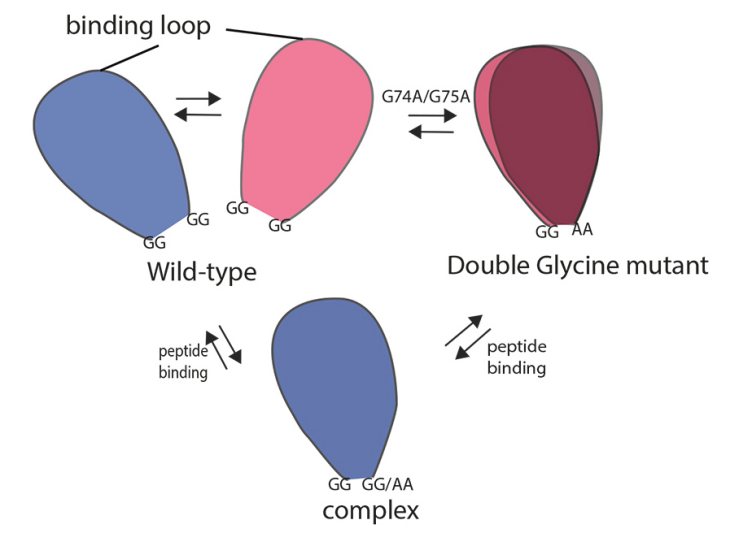
NMR is one of the principal experimental techniques in structural biology with abilities to determine atomic resolution structures as well as to investigate dynamic features and intermolecular interactions of biological macromolecules with a molecular weight smaller than 30 kDa. With the techniques TROSY and CRIPT, NMR is the multiprobe method to study protein-protein or protein-drug interactions of structures with a molecular weight up to 1 MDa. With the technique exact NOEs (eNOEs) we established a method to determine distances within a protein within an accuracy of 0.1 Å allowing the determination of a structural ensemble reflecting the various states of a protein. The application of eNOEs to protein-protein interaction and enzymes opens an avenue towards determining the dynamics of a protein at atomic resolution and thus permitting the structure and dynamics activity relationships of proteins in action.